
In the last few years a multitude of new reconstruction approaches have been proposed in diffusion MRI, each group testing the performances of their proposed algorithms with their own synthetic data and evaluation criteria. When proposing a new approach, then, it can be really hard to compare the performances against the state-of-the-art techniques.
The diffusion MRI reconstruction contest is organized with the aim to provide all researchers in this field with a common framework to test their own algorithms on given synthetic datasets and fairly compare their results against other approaches, thus paving the way for a common way to assess the performances and evaluate the results of each method.
The registration procedure is very simple: just send an email to the organizers with the following details:
The registration is open to everyone.
Each team participating to the contest will be requested to write a 1-page abstract describing the approach undertaken. All papers will be gathered to form the workshop proceedings. These proceedings will be published on this website and on the official IEEE International Symposium on Biomedical Imaging (ISBI 2012) website.
Please refer to this template for typesetting your abstract. Since the results will be unknown at the time of submission of the abstract, only a technical description of the method is indeed requested.
The purpose of the contest is to compare different reconstruction approaches on the same synthetic data and under controlled conditions. To do this, some training data will be released prior of the contest itself, thus allowing the contestants to practice with the data format and test their methods on it. To draw the final ranking, however, another dataset will be used, but this time the ground-truth will be unknown to the contestants.
Note
Contest details might be subjected to changes before the release of the testing dataset. We are receiving some very valuable comments and suggestions about the framework of the contest and we have reserved ourselves the rights to include them for the final contest in order to be as fair as possible. However, the final rules will be published before the testing data is released and will not be modified any further.
As mentioned above, before the real contest we will release a training data phantom to permit the participants to practice and test their methods with the same kind of data they will be evaluated later on for the final ranking of the contest. This data can be download from the (download section). The ground-truth of this synthetic dataset will be disclosed, meaning that the fibers configuration in each voxel will be revealed to the contestants.
The data will be accompanied with the MATLAB code to simulate the signal at any position in q-space. The participants can simulate the signal corresponding to whatever acquisition scheme they need, without limitations on the number or positions of the samples. They can do whatever they want on this data, there are no limitation on the number of samples to simulate, the experiments they can perform or the algorithms they use. Basically, there are no rules here!
Concerning the testing data, which is the synthetic phantom used to evaluate each method and to draw the final ranking of the contest, the ground-truth will be, of course, unknown to the participants. The fibers configuration in each voxel of this dataset will be disclosed only when the final ranking of the contest are published. The format of the data is the same as for the testing data; only the configuration of the fiber compartments in each voxel changes.
Important
The ground-truth of the testing data will be unknown to the contestants! The actual fibers configuration in each voxel of this dataset will be revealed when the final ranking is published.
In order to probe the signal, the contestants will submit to the organizers their acquisition scheme (i.e. one “gradient_list.txt” file) with the q-space positions to probe. Then, they will receive back a file with the signal simulated at each requested sampling point and with different level of noise. In particular, the performances of each method with data having an SNR of 10, 20 and 30 will be tested. At this point, each team can apply his proposed reconstruction method on these signal samples and will then submit to the organizers the estimated fibers configuration in each voxel.
The evaluations of the performances of each method and the final ranking will be published on this website together with the ground-truth of the testing dataset.
Important
Each participant can request the organizers only one acquisition scheme (i.e. “gradient_list.txt”) containing the q-space coordinates where to simulate the signal corresponding to the testing data phantom.
Every synthetic phantom used throughout the contest (both for training and testing) is actually made of two distinct voxel fields:
Isolated voxels dataset.
This dataset consists in a matrix where each voxel has no spatial relations with its neighbours, so they can be considered independent one another and there isn’t any structure in this dataset. Each voxel contains a random fibers configuration (i.e. the number of fibers can change, their orientations etc). In particular, the dataset will contain voxels with different combinations of:
- number of fiber compartments;
- directions of the main axis of each fiber compartment;
- diffusivity profile (diffusivities).
The purpose of this dataset is to assess the average performance of each reconstruction method in disentangling distinct fiber compartments under specific experimental conditions.
Structured field dataset.
This datasets aims to simulate more closely a “real” fibers configuration occuring in reality; several “fiber bundles” are generated having spatial coherence of diffusion properties across neighbouring voxels (see the following figure). The purpose of this dataset is to test the ability of each proposed method to exploit information from neighboring voxels during the reconstruction of the fiber configuration at a given position.
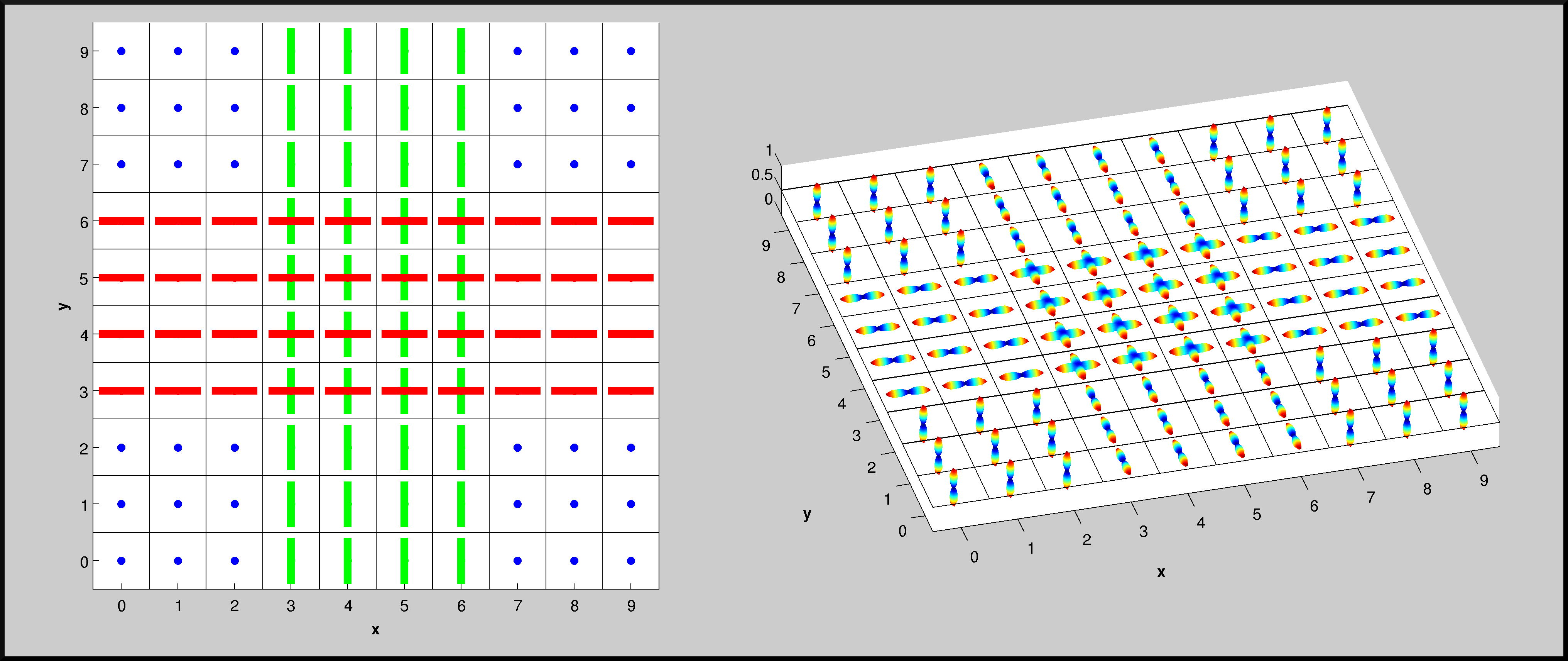
Screenshots of a possible synthetic dataset used during the contest. On the left panel, the directions of the fiber compartments in each voxel are reported; as usual, they are color-coded depending on their direction (RED: x-axis, GREEN: y-axis, BLUE: z-axis). On the right panel, instead, the ODFs are plotted for each voxel; ODFs are analytically computed from the model.
In the training phase there are actually no rules! Every participant can probe the signal corresponding to the training data for an unlimited number of times and for every combination of sampling schemes. The structure of this synthetic phantom (i.e. number of fiber compartments in each voxel, orientation etc) will be known to the teams.
Concerning the testing phase some rules apply; here it is a brief summary:
Two main criteria will be considered for determining the final ranking: the reconstruction quality and the number of samples probed. As a general rule, the method achieving the best reconstruction quality with the smallest number of samples will win the contest. Of course the balance among these two criteria is very challenging. We are putting all our might in order to come up with a “fair” trade-off, but we are very open to suggestions in this respect.
At the voxel level, a score will be attributed to each reconstruction method accounting for the following metrics:
Correct estimation of the number of fiber compartments, expressed by means of the probability of false fibre detection:
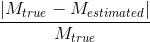
where 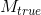 and
and 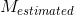 are, respectively, the real and estimated number of fiber compartments inside the voxel.
are, respectively, the real and estimated number of fiber compartments inside the voxel.
Angular precision of the estimated fiber compartments, assessed by means of the angular error (in degrees) between the estimated fiber directions and the true ones inside the voxel:
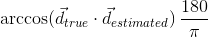
where 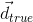 is a true direction inside the voxel and
is a true direction inside the voxel and 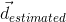 is its closest estimate. The final value will be the average of the angular errors computed for all the true fiber compartments. Missing and additional fiber compartments will be penalyzed proportionally to the corresponding volume fraction.
is its closest estimate. The final value will be the average of the angular errors computed for all the true fiber compartments. Missing and additional fiber compartments will be penalyzed proportionally to the corresponding volume fraction.
Accuracy in the estimation of the ODF, computed as the normalized mean squared error between the estimated ODF, 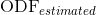 , and the one analytically computed from the model used for the simulations,
, and the one analytically computed from the model used for the simulations, 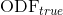 :
:
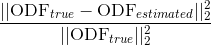
Distinct rankings will be created for every metric, different levels of noise and given specific conditions (i.e. crossing angles). Points will be assigned to each method based on the position of each standing (e.g. 10 points to the 1st, 6 to the 2nd etc.) The reconstruction method accumulating the highest number of points will win the contest!
Note
More details about the exact evaluation of the performances of each reconstruction method (i.e. exact computation of the scores and final ranking) will be published only after the results have been submitted. We do no want to force groups to “tweak” and “tune” their methods depending on the exact evaluation criteria. We would like to fairly compare the methods with the standard metrics used in the literature (e.g. ([Michailovic2011], [Landman2011])). However, as usual, we are open to suggestions and discussions. Should you have any comments, please do not hesitate to contact us by due time!
In this page we list all the registered teams:
| Team | Members | Method |
|---|---|---|
| Tocororo01 | Erick Jorge Canales-Rodríguez, Lester Melie-García, Yasser Iturria-Medina, Yasser Alemán-Gómez CIBERSAM and FIDMAG Hermanas Hospitalarias Benito Menni CASM (Barcelona, Spain), Cuban Neuroscience Center (Havana, Cuba), Department of Experimental Medicine and Surgery, Hospital General Universitario Gregorio Marañón (Madrid, Spain) |
Reconstruction method described in (E.J. Canales-Rodríguez, C.P. Lin, Y. Iturria-Medina, C.H. Yeh, K.H. Cho and L. Melie-García, “Diffusion orientation transform revisited”, Neuroimage, Jan 15;49(2):1326-39, 2010) |
| Tocororo02 | Erick Jorge Canales-Rodríguez, Lester Melie-García, Yasser Iturria-Medina, Yasser Alemán-Gómez CIBERSAM and FIDMAG Hermanas Hospitalarias Benito Menni CASM (Barcelona, Spain), Cuban Neuroscience Center (Havana, Cuba), Department of Experimental Medicine and Surgery, Hospital General Universitario Gregorio Marañón (Madrid, Spain) |
Reconstruction method described in (E.J. Canales-Rodríguez, Y. Iturria-Medina, Y. Alemán-Gómez and L. Melie-García, “Deconvolution in diffusion spectrum imaging”, Neuroimage, Mar;50(1):136-49, 2010) |
| The HARDY | Farshid Sepehrband, Jayran Chupan, Quang Tieng, Viktor Vegh, Steven Yang Centre for Advanced Imaging, The University of Queensland (Brisbane, Australia) |
Iterative scheme that employs super-resolution techniques similar to track-density imaging to create tractography at higher anatomical resolution, which in turn is used as prior knowledge to regularize the HARDI reconstruction |
| The HOT gang | Yaniv Gur SCI Institute, University of Utah (Salt Lake City, USA) |
Low-rank approximations of Higher Order Tensors (HOT) |
| MrSCIL | Michael Paquette, Maxime Descoteaux Sherbrooke Connectivty Imaging Lab, Sherbrooke University (Sherbrooke, Quebec, Canada) |
Method under development |
| MrSS | Maxime Descoteaux, Arnaud Boré Sherbrooke Connectivty Imaging Lab, Sherbrooke University (Sherbrooke, Quebec, Canada) |
Single-shell HARDI reconstruction method |
| NIPG | Ying-Chia Lin, Gloria Menegaz University of Verona (Verona, Italy) |
Multi-resolution denoising approach |
| Athena | Emmanuel Caruyer, Sylvain Merlet, Aurobrata Ghosh, Rachid Deriche Inria (Sophia-Antipolis Méditerranée, France) |
Method under development |
| Frogs | Alonso Ramirez-Manzanares, Angel Ramon Aranda Campos, Mariano Rivera Meraz University of Guanajuato (Guanajuato, Mexico), Research Center on Mathematics, CIMAT, A.C., (Guanajuato, Mexico) |
Improved version of the Diffusion Basis Functions Method (A. Ramírez-Manzanares, M. Rivera, et al, “Diffusion basis functions decomposition for estimating white matter intravoxel fiber geometry”, IEEE TMI, 26(8), 2007) |
| Hawks | Merry Mani, Mathews Jacob, Jianhui Zhong University of Rochester (Rochester, USA), University of Iowa (Iowa City, USA) |
Method under development |
| The Diffusiontamers | Paulo Rodrigues, Vesna Prčkovska Universitat de Barcelona (Barcelona, Spain), IDIBAPS (Barcelona, Spain) |
Method under development |
| Tyranny | Eleftherios Garyfallidis University of Cambridge (Cambridge, United Kingdom) |
The Equatorial Inversion Transform |
| Frunik | Marco Reisert, Valerij Kiselev, Henrik Skibbe Department of Radiology, University Medical Center (Freiburg, Germany) |
L1-based deconvolution with spatial regularization |
This section reports the results of the preliminary analysis performed at the time of the challenge. Please note that some results are missing (not available at the time ot the submission) or corrupted (issues in the data format like orientation problem etc).
Please refer to the latest analysis for more recent and detailed results.
This section reports the individual performace of each team.
In this section we report the comparison among groups.
As discussed during the workshop, finding the proper trade-off between the quality a methoid achieves and the number of samples it uses was very though, if not impossible at all to estimate since it depends a lot on the question one wants to answer.
Anyway, for the scope of this contest, three different weighting functions 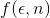 have been proposed:
have been proposed:
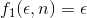
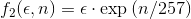
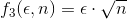
where  and
and 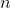 are, respectively, the value of one of the given error metrics and the number of samples used.
are, respectively, the value of one of the given error metrics and the number of samples used.
Of course, increasing the weighting degree has the effect of favoring methods with less points. The weighting function has been democratically selected among the participants by email before revealing the comparison results. The function 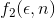 was the chosen one.
was the chosen one.
In the following, we report the three separate standings created by using the three aforementioned weighting functions.
References
| [Michailovic2011] | Michailovic et al. Spatially regularized compressed sensing for high angular resolution diffusion imaging. IEEE TMI, 30: 1100-1115 (2011) |
| [Landman2011] | Landman et al. Resolution of crossing fibers with constrained compressed sensing using diffusion tensor MRI. Neuroimage, In press (2011) |