Testing data¶
DWI images have been generated for predetemined sampling schemes, accordingly to the sampling classes of the contest (see presentation of sampling classes).
If you have an optimized sequence or specific needs for the acquisition, you are encouraged to request additional diffusion weighted images of the same testing data. In order to simulate the signal corresponding to the testing dataset, please send an email to the organizers with the acquisition scheme containing the q-space coordinates to probe. This should be described in a .bvec and .bvec text files. The signal will be simulated at each requested sampling point as described in the Signal Simulation section and using different levels of noise (SNR = 10, 20, 30).
At this point, each team can apply his proposed reconstruction method on the received signal and will then submit to the organizers the estimated fibers configuration in each voxel of the testing dataset (see Results Submission for details) for the final evaluation.
The evaluations of the performance of each method and the final ranking will be published on this website, together with the ground-truth of the testing dataset, only the day of the workshop (see Ground-truth orientations section).
Rules¶
- Each participant can request only one acquisition scheme.
- Data sharing among teams is forbidden.
- Merging data coming from acquisitions at different SNRs is forbidden. The reconstructions estimated for a datset with a given SNR must be computed using exclusively the data at that SNR.
- Results must be reproducible.
Gradient list¶
The gradient directions and b-values should be stored in two separate text files:
- one file with extension .bval. This contains, on a single row, b-values for all the sampling directions, separated by a space. See example of a .bval file.
- one file with extension .bvec. This contains, on three sepoarate rows, the x, y and z coordinates (respectively) of the unit gradient directions. See example of a .bvec.
Please make sure the gradient list you request the signal for corresponds to the appropriate sampling class (see Sampling classes section). For instance, a gradient scheme wiith 65 gradient directions will be considered as a Heavyweight sampling, and evaluated with methods such as DSI.
Submitting the results¶
Each participant should return to the organizers the fibers configuration in each voxel of the testing dataset as estimated with their reconstruction technique. One separate file is requested for each SNR.
One has two options to submit results:
A spherical harmonics (SH) representation of your estimated ODF, odf_sh.nii. As there is a plethora of existing bases functions, we require the following:
- The SH basis used must follow the MRTRIX basis convention. We provide an example showing how to generate such a SH representation using the python script, dipy/bin/dipy_sh_estimate.py.
- Maximum SH order of r=16. Let R = (r+1)(r+2)/2, the final file should thus be a 50x50x50xR nifti file, encoded on 32 bits floats.
A peak file, peaks.nii. We require the following:
- Maximum of 5 peaks per voxel, p1, p2, ..., p5. We provide an example showing how to generate such a peak file using the python script, dipy/bin/dipy_peak_extraction.py.
- Each peak is represented in Cartesian coordinates pi = (xi, yi, zi) with its norm proportional to its fraction in the voxel. pi = (0,0,0) if there is no peak.
- The final file should thus be a 50x50x50x15 nifti file, encoded on 32 bits floats.
Important
Every team must make sure that there odf_sh.nii file or peaks.nii file loads in the FiberNavigator to make sure there is no flipping problem. A YouTube video tutorial is available to help you.
Technical details¶
Here are some technical directions so as to smoothly submit the results.
You will be transferring large files. We recommend the use of a solution like https://www.wetransfer.com/ to avoid sending these files by email.
To communicate the organizers the link of the web service hosting your submissions (or ftp servers in your home institution), please use the following email address: tractometer@gmail.com.
We would greatly appreciate if you could format your file name with the strict following conventions:
- all lower-case characters, allowed characters are a-z, 0-9, -, _.
- format file names as follows: <name-of-team>_<sampling-category>_<peaks/odfs>_snr-<SNR>.nii.gz
Examples of file names:
john-smith_dti_peaks_snr-20.nii.gz
the-dream-team_hardi_odfs_snr-10.nii.gz
Thanks for your cooperation.
Checking your reconstructions¶
This section shows two ways for visually inspecting your reconstructions, to spot quickly any possible problem in the data, e.g. flipping or swapping of axes.
With Dipy¶
To run these scripts, you will need to install Dipy. See the documentation for more information.
Important
For a proper execution of these scripts, you must use cython 0.17+ and the latest dipy-0.6-dev from github.
Once Dipy is installed and configured, you can execute the scripts this way:
$ python dipy/bin/dipy_sh_estimate.py odf.nii.gz sphere724.txt odf_sh.nii.gz
$ python dipy/bin/dipy_peak_extraction.py odf.nii.gz sphere724.txt peaks.nii.gz
$ python viz_peaks.py
Files provided:
- gfa.nii.gz (generalized fractional anisotropy map)
- odf.nii.gz (orientation distribution function on the sphere with 724 uniform points)
- sphere724.txt (sphere with 724 points)
- viz_peaks.py
Important
Please note that in the script dipy_sh_estimate.py the regularization weight 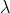 is set to 0.006 by default.
If you wish to tune the regularization weight, an additional parameter -l is available, e.g. use -l 0.0 if you do not want to regularize at all during the fitting of the SH coefficients.
is set to 0.006 by default.
If you wish to tune the regularization weight, an additional parameter -l is available, e.g. use -l 0.0 if you do not want to regularize at all during the fitting of the SH coefficients.
Documentation¶
Dipy:
FiberNavigator:
- http://code.google.com/p/fibernavigator/downloads/list (Windows and OSX packages)
- http://code.google.com/p/fibernavigator/wiki/UbuntuBuildInstructions (Build instructions for Ubuntu)
- tutorial for loading ODF/peaks into the FiberNavigator
Classical sampling schemes¶
| Description | Sampling scheme | Diffusion-weighted images |
|---|---|---|
| 32 directions, b = 1200 s/mm² | ||
| 64 directions, b = 3000 s/mm² | ||
| 515 acquisitions, b < 12000 s/mm² |
Ground-truth orientations¶
To simplify the self-assessment of the quality of own reconstructions, we provide a script to compute standard quality metrics in each voxel (compute_local_metrics.py) together with the file of the ground-truth peaks (ground-truth-peaks.nii.gz):